16S rRNA Metagenomics Sequencing
Metagenomic sequencing is a technique used to sequence the DNA extracted from all organisms (typically bacteria and fungi) present in a bulk sample. It can be used to study specific communities of microorganisms, such as those on the skin, in soil or in a water sample. Typically, short highly conserved genes or intergenic regions are targeted with specific PCR primers and analysed using next generation sequencing (NGS), making it possible to analyse them in a wide range of different microorganisms in a single sample. These regions include 16S rRNA and the ITS.
ITS
The Internal transcribed spacer (ITS) is a spacer DNA located between the small-subunit ribosomal RNA (rRNA) and large-subunit rRNA genes in the chromosome. It is the most widely sequenced region for those interested in fungal ecology and has been recommended as the universal fungal barcode sequence. This allows it to be used to assess the diversity of the fungal population within environmental or clinical samples.
For metagenomic sequencing, we use the Illumina MiSeq system along with appropriate flow cells and consumables for your required read depth. Our bioinformaticians use dedicated analysis software to map your data to bacterial and fungal genome databases and can assess microbial richness and diversity using the abundance-based coverage estimator (ACE), Chao1, Shannon or Simpson indices.
16S RNA
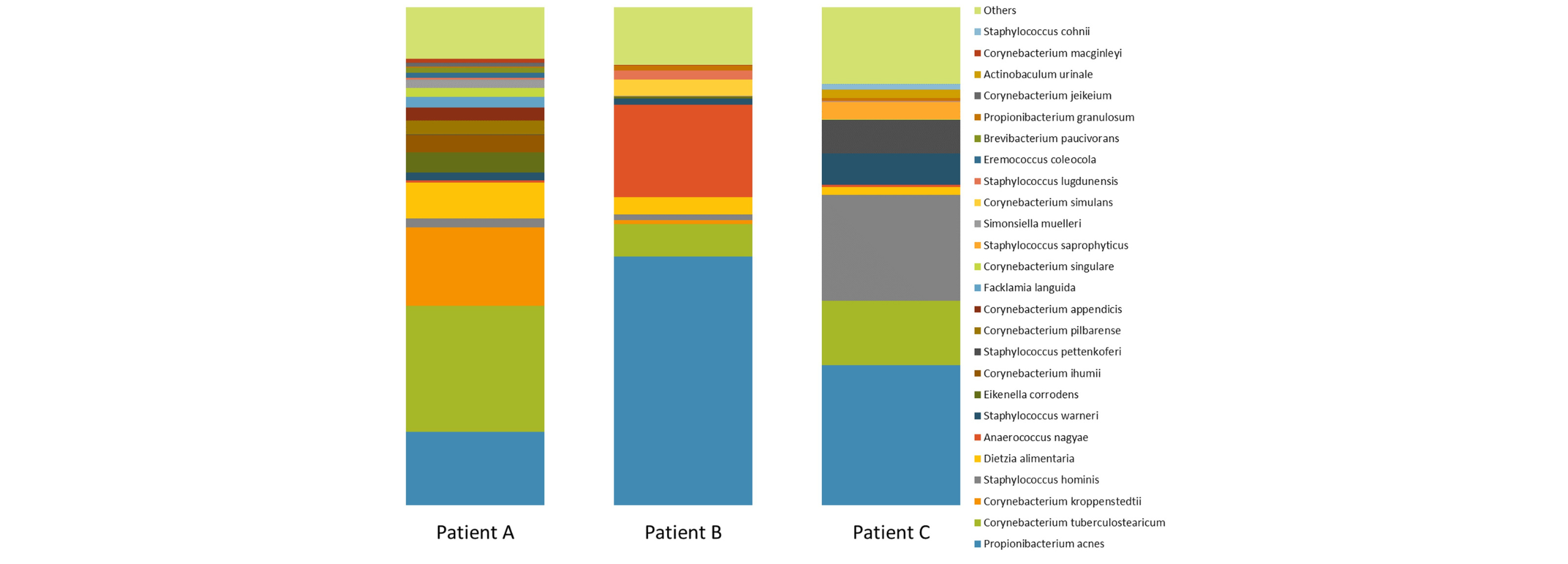
16S ribosomal RNA (16S rRNA) is a component of the prokaryotic ribosome, which acts as a scaffold helping define the positions of ribosomal proteins. It contains nine hypervariable regions that can provide species-specific signature sequences useful for the identification of bacteria. There are highly conserved sequences between these hypervariable regions which allow us to discriminate between specific microorganisms such as bacteria, archaea and eukarya within a bulk sample.
Metatranscriptomics
While metagenomics focuses on identifying which microbes are present within a community, metatranscriptomics can be used to study the diversity of the active genes within the community, to quantify their expression levels and to monitor how these levels change in different conditions. Metatranscriptomics can provide information about differences in the active functions of microbial communities that would otherwise appear to have a similar make-up.
We use Illumina Stranded Total RNA Prep with Ribo-Zero Plus Microbiome, which effectively depletes unwanted host RNA and bacterial rRNA from the sample to allow for high-quality metatranscriptomics assessment.